HOME  Research Projects
Research Projects  Gut Design Research Project
Gut Design Research Project
Gut Design Research Project
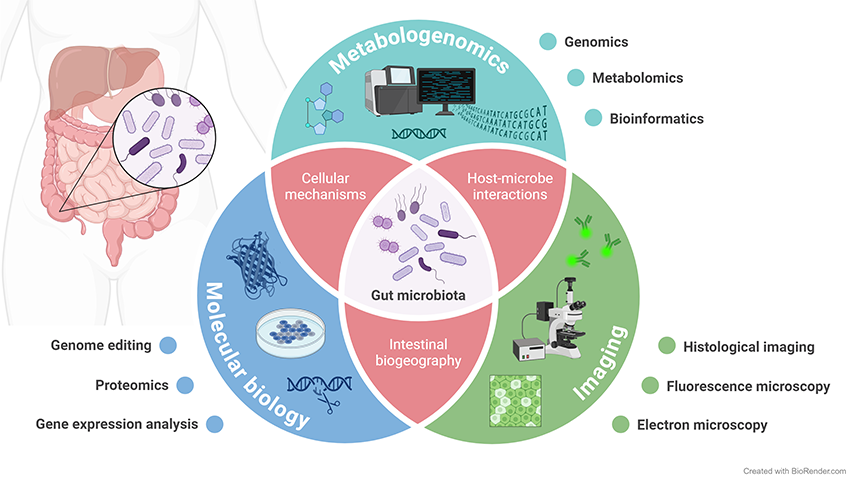
The Gut Design Research Project aims to elucidate the relationship between gut microbiota and human health, focusing on methods to regulate the microbiota effectively (Gut Design). Our intestines host diverse microorganisms that play essential roles, such as supporting the immune system and providing vital nutrients. However, an imbalance in these communities can lead to the onset or worsening of various diseases. By uncovering the mechanisms of symbiosis between gut microbiota and their host, we aim to establish innovative approaches for health management and disease prevention or treatment. This project employs a multi-omics approach, combining advanced techniques such as microbiota structural analysis, metabolome profiling, bioinformatics, immunological analysis, and bacterial culture technology. We believe that a comprehensive understanding of gut microbiome functions, still a largely uncharted field, is the first step toward achieving effective gut design.
Publication
- Steimle A, Neumann M, Grant ET, Willieme S, De Sciscio A, Parrish A, Ollert M, Miyauchi E, Soga T, Fukuda S, Ohno H, Desai MS. Gut microbial factors predict disease severity in a mouse model of multiple sclerosis. Nat. Microbiol. 9: 2244, 2024.
- Song I, Yang J, Saito M, Hartanto T, Nakayama Y, Ichinohe T, Fukuda S*. Prebiotic inulin ameliorates SARS-CoV-2 infection in hamsters by modulating the gut microbiome. npj Sci. Food. 8: 18, 2024.
- Tsukimi T, Obana N, Shigemori S, Arakawa K, Miyauchi E, Yang J, Song I, Ashino Y, Wakayama M, Soga T, Tomita M, Ohno H, Mori H, Fukuda S*. Genetic mutation in Escherichia coli genome during adaptation to the murine intestine is optimized for the host diet. mSystems. 9: e0112323, 2024.
- Nagai M, Moriyama M, Ishii C, Mori H, Watanabe H, Nakahara T, Yamada T, Ishikawa D, Ishikawa T, Hirayama A, Kimura I, Nagahara A, Naito T*, Fukuda S*, Ichinohe T*. High body temperature increases gut microbiota-dependent host resistance to influenza A virus and SARS-CoV-2 infection. Nat. Commun. 14: 3863, 2023.
- Kawamoto S, Uemura K, Hori N, Takayasu L, Konishi Y, Katoh K, Matsumoto T, Suzuki M, Sakai Y, Matsudaira T, Adachi T, Ohtani N, Standley DM, Suda W, Fukuda S, Hara E. Bacterial induction of B cell senescence promotes age-related changes in the gut microbiota. Nat. Cell Biol. 25: 865, 2023.
- Zhu X, Sakamoto S, Ishii C, Smith MD, Ito K, Obayashi M, Unger L, Hasegawa Y, Kurokawa S, Kishimoto T, Li H, Hatano S, Wang TH, Yoshikai Y, Kano SI, Fukuda S, Sanada K, Calabresi PA, Kamiya A. Dectin-1 signaling on colonic γδ T cells promotes psychosocial stress responses. Nat. Immunol. 24: 625, 2023.
- Morita H, Kano C, Ishii C, Kagata N, Ishikawa T, Hirayama A, Uchiyama Y, Hara S, Nakamura T, Fukuda S*. Bacteroides uniformis and its preferred substrate, α-cyclodextrin, enhances endurance exercise performance in mice and human males. Sci. Adv. 9: eadd2120, 2023.
- Yang J, Tsukimi T, Yoshikawa M, Suzuki K, Takeda T, Tomita M, Fukuda S*. Cutibacterium acnes (Propionibacterium acnes) 16S rRNA genotyping of microbial samples from possessions contributes to owner identification. mSystems 4: e00594-19, 2019.
- Yachida, S., Mizukami, S., Shiroma, H., Shiba, S., Nakajima, T., Sakamoto, T., Watanabe, H., Masuda, K., Nishimoto, Y., Kubo, M., Hosoda, F., Rokutan, H., Matsumoto, M., Takamaru, H., Yamada, M., Matsuda, T., Iwasaki, M., Yamaji, T., Yachida, T., Soga, T., Kurokawa, K., Toyoda, A., Ogura, Y., Hayashi, T., Hatakeyama, M., Nakagama, H., Saito, Y., Fukuda, S., Shibata, T., Yamada, T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25: 968-976, 2019.
- Ishii, C., Nakanishi, Y., Murakami, S., Nozu, R., Ueno, M., Hioki, K., Aw, W., Hirayama, A., Soga, T., Ito, M., Tomita, M., *Fukuda, S. A metabologenomic approach reveals changes in the intestinal environment of mice fed on American diet. Int. J. Mol. Sci. 19: E4079, 2018.

Shinji Fukuda
Project Professor

Jiayue Yang
Project Research Associate

Wanping AW
Project Research Associate

Isaiah Youhak Song
Project Research Associate

Chiharu Ishii
Visiting Research Fellow
- List of research projects
- Postembryonic development in crustaceans
- Molecular Anhydrobiology Project
- Protein Materials Project
- Food metabolomics project
- Functional RNA Analysis Project
- 3D Cell Atlas Project
- Bio-Functional Design Project
- Molecular Oncology Project
- Medical Informatics Project
- Metabolomics Project
- Bioenergetic regulation Project
- Cancer Metabolism Project
- Tsuruoka Metabolomics Cohort Study (TMCS) Project
- Environmental systems biology Project
- Drug delivery system project
- Extracellular vesicle molecular function research Project
- Gut Design Research Project
- Kleptobiolgy Project
- Bacterial regulatory RNA project
- Synthetic Biology Project
- Ribosome & Proteomics Project